EF手
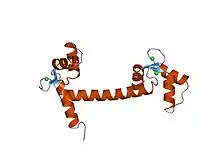
EF手包括一个螺旋-环-螺旋拓扑结构,就像人类的手上拇指和食指,而Ca2+离子与其配体就位于环内。这一结构最早在小清蛋白中发现,小清蛋白含有三个EF手结构,与钙离子结合而造成的肌肉放松有关。
| EF手 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| 鑑定 | |||||||||||
| 標誌 | efhand | ||||||||||
| Pfam | PF00036 | ||||||||||
| InterPro | IPR002048 | ||||||||||
| PROSITE | PDOC00018 | ||||||||||
| SCOP | 1osa / SUPFAM | ||||||||||
| OPM蛋白 | 1djx | ||||||||||
| |||||||||||
EF手(英語:)是一种在钙结合蛋白中发现的螺旋-环-螺旋结构域或结构基序。
EF手包含两个α螺旋,由一个短的(通常包括12个氨基酸)环状区域连接,常用于结合钙离子。EF手也出现在钙调蛋白的所有结构域和肌肉蛋白质肌钙蛋白C中。
钙离子结合位点
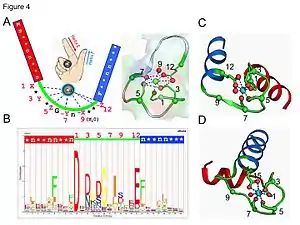
EF-hand Ca2+ binding motif.
- The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X, Y, Z, -Y, -X and -Z. The invariant Glu or Asp at position 12 provides two oxygens for liganding Ca (bidentate ligand).
- The calcium ion is bound by both protein backbone atoms and by amino acid side chains, specifically those of the acidic amino acid residues aspartate and glutamate. These residues are negatively charged and will make a charge-interaction with the positively charged calcium ion. The EF hand motif was among the first structural motifs whose sequence requirements were analyzed in detail. Five of the loop residues bind calcium and thus have a strong preference for oxygen-containing side chains, especially aspartate and glutamate. The sixth residue in the loop is necessarily glycine due to the conformational requirements of the backbone. The remaining residues are typically hydrophobic and form a hydrophobic core that binds and stabilizes the two helices.
- Upon binding to Ca2+, this motif may undergo conformational changes that enable Ca2+-regulated functions as seen in Ca2+ effectors such as calmodulin (CaM) and troponin C (TnC) and Ca2+ buffers such as calreticulin and calbindin D9k. While the majority of the known EF-hand Calcium-binding proteins (CaBPs) contain paired EF-hand motifs, CaBP’s with single EF hands have also been discovered in both bacteria and eukaryotes. In addition, “EF-hand-like motifs” have been found in a number of bacteria. Although the coordination properties remain similar with the canonical 29-residue helix-loop-helix EF-hand motif, the EF-hand-like motifs differ from EF-hands in that they contain deviations in the secondary structure of the flanking sequences and/or variation in the length of the Ca2+-coordinating loop.
Prediction

Summary of motif signatures used for prediction of EF-hands.
- Pattern (motif signature) search is one of the most straightforward ways to predict continuous EF-hand Ca2+-binding sites in proteins. Based on the sequence alignment results of canonical EF-hand motifs, especially the conserved side chains directly involved in Ca2+ binding, a pattern Template:PROSITE has been generated to predict canonical EF-hand sites. A prediction servers may be found in the external links section.
分类
- 自从EF手结构模体在1973年被描绘以来,迄今为止该家族的蛋白质已扩展到至少66个亚家族。EF手结构基元可以分为两大类:
- Canonical EF-hands as seen in calmodulin (CaM) and the prokaryotic CaM-like protein calerythrin. The 12-residue canonical EF-hand loop binds Ca2+ mainly via sidechain carboxylates or carbonyls (loop sequence positions 1, 3, 5, 12). The residue at the –X axis coordinates the Ca2+ ion through a bridged water molecule. The EF-hand loop has a bidentate ligand (Glu or Asp) at axis –Z.
- Pseudo EF-hands exclusively found in the N-termini of S100 and S100-like proteins. The 14-residue pseudo EF-hand loop chelates Ca2+ primarily via backbone carbonyls (positions 1, 4, 6, 9).
Additional points:
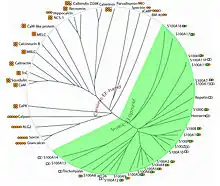
EF手蛋白家族的系统发育树
- EF-hand-like proteins with diversified flanking structural elements around the Ca2+-binding loop have been reported in bacteria and viruses. These prokaryotic EF-hand-like proteins are widely implicated in Ca2+ signaling and homeostasis in bacteria. They contain flexible lengths of Ca2+-binding loops that differ from the EF-hand motifs. However, their coordination properties resemble classical EF-hand motifs.
- For example, the semi-continuous Ca2+-binding site in D-galactose-binding protein (GBP) contains a nine-residue loop. The Ca2+ ion is coordinated by seven protein oxygen atoms, five of which are from the loop mimicking the canonical EF-loop whereas the other two are from the carboxylate group of a distant Glu.
- Another example is a novel domain named Excalibur (extracellular Ca2+-binding region) isolated from Bacillus subtilis. This domain has a conserved 10-residue Ca2+-binding loop strikingly similar to the canonical 12-residue EF-hand loop.
- The diversity of the structure of the flanking region is illustrated by the discovery of EF-hand-like domains in bacterial proteins. For example, a helix-loop-strand instead of the helix-loop-helix structure is in periplasmic galactose-binding protein (Salmonella typhimurium, PDB 1gcg) or alginate-binding protein (Sphingomonas sp., 1kwh); the entering helix is missing in protective antigen (Bacillus anthracis, 1acc) or dockerin (Clostridium thermocellum, 1daq).
- Among all the structures reported to date, the majority of EF-hand motifs are paired either between two canonical or one pseudo and one canonical motifs. For proteins with odd numbers of EF-hands, such as the penta-EF-hand calpain, EF-hand motifs were coupled through homo- or hetero-dimerization. The recently-identified EF-hand containing ER Ca2+ sensor protein, stromal interaction molecule 1 and 2 (STIM1, STIM2), has been shown to contain a Ca2+-binding canonical EF-hand motif that pairs with an immediate, downstream atypical "hidden" non-Ca2+-binding EF-hand. Single EF-hand motifs can serve as protein-docking modules: for example, the single EF hand in the NKD1 and NKD2 proteins binds the Dishevelled (DVL1, DVL2, DVL3) proteins.
- Functionally, the EF-hands can be divided into two classes: 1) signaling proteins and 2) buffering/transport proteins. The first group is the largest and includes the most well-known members of the family such as calmodulin, troponin C and S100B. These proteins typically undergo a calcium-dependent conformational change which opens a target binding site. The latter group is represented by calbindin D9k and do not undergo calcium dependent conformational changes.
亚族
- EPS15 homology (EH) domain – IPR000261
例子
Humans proteins containing this domain include:
- ACTN1、ACTN2、ACTN3、ACTN4、APBA2BP; AYTL1; AYTL2
- C14orf143; CABP1; CABP2; CABP3; CABP4; CABP5; CABP7; CALB1; CALB2; CALM2; CALM3; CALML3; CALML4; CALML5; CALML6; CALN1; CALU; CAPN1; CAPN11; CAPN2; CAPN3; CAPN9; CAPNS1; CAPNS2; CAPS; CAPS2; CAPSL; CBARA1; CETN1; CETN2; CETN3; CHP; CHP2; CIB1; CIB2; CIB3; CIB4; CRNN
- DGKA; DGKB; DGKG; DST; DUOX1; DUOX2
- EFCAB1; EFCAB2; EFCAB4A; EFCAB4B; EFCAB6; EFCBP1; EFCBP2; EFHA1; EFHA2; EFHB; EFHC1; EFHD1; EFHD2; EPS15; EPS15L1
- FKBP10; FKBP14; FKBP7; FKBP9; FKBP9L; FREQ; FSTL1; FSTL5
- GCA; GPD2; GUCA1A; GUCA1B; GUCA1C
- hippocalcin; HPCAL1; HPCAL4; HZGJ
- IFPS; ITSN1; ITSN2; KCNIP1; KCNIP2; KCNIP3; KCNIP4; KIAA1799
- LCP1
- MACF1; MRLC2; MRLC3; MST133; MYL1; MYL2; MYL5; MYL6B; MYL7; MYL9; MYLC2PL; MYLPF
- NCALD; NIN; NKD1; NKD2; NLP; NOX5; NUCB1; NUCB2
- OCM
- PDCD6; PEF1; PKD2; PLCD1; PLCD4; PLCH1; PLCH2; PLS1; PLS3; PP1187; PPEF1; PPEF2; PPP3R1; PPP3R2; PRKCSH; PVALB
- RAB11FIP3; RASEF; RASGRP; RASGRP1; RASGRP2; RASGRP3; RCN1; RCN2; RCN3; RCV1; RCVRN; REPS1; RHBDL3; RHOT1; RHOT2; RPTN; RYR2; RYR3
- S100A1; S100A11; S100A12; S100A6; S100A8; S100A9; S100B; S100G; S100Z; SCAMC-2; SCGN; SCN5A; SDF4; SLC25A12; SLC25A13; SLC25A23; SLC25A24; SLC25A25; SPATA21; SPTA1; SPTAN1; SRI
- TBC1D9; TBC1D9B; TCHH; TESC; TNNC1; TNNC2
- USP32
- VSNL1
- ZZEF1
另见
- 锚定结构域,另一种由α-螺旋组成的钙结合结构基序。
参考文献
- Ban C, Ramakrishnan B, Ling KY, Kung C, Sundaralingam M. . Acta Crystallogr. D Biol. Crystallogr. January 1994, 50 (Pt 1): 50–63. PMID 15299476. doi:10.1107/S0907444993007991.
延伸阅读
- Branden C, Tooze J. . . New York: Garland Pub. 1999: 24–25. ISBN 0-8153-2305-0.
- Nakayama S, Kretsinger RH. . Annu Rev Biophys Biomol Struct. 1994, 23: 473–507. PMID 7919790. doi:10.1146/annurev.bb.23.060194.002353.
- Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ. . Proteins. November 2006, 65 (3): 643–55. PMID 16981205. doi:10.1002/prot.21139.
- Zhou Y, Frey TK, Yang JJ. . Cell Calcium. July 2009, 46 (1): 1–17. PMID 19535138. doi:10.1016/j.ceca.2009.05.005.
- Nakayama S, Moncrief ND, Kretsinger RH. . J. Mol. Evol. May 1992, 34 (5): 416–48. PMID 1602495. doi:10.1007/BF00162998.
- Hogue CW, MacManus JP, Banville D, Szabo AG. . J. Biol. Chem. July 1992, 267 (19): 13340–7. PMID 1618836.
- Bairoch A, Cox JA. . FEBS Lett. September 1990, 269 (2): 454–6. PMID 2401372. doi:10.1016/0014-5793(90)81214-9.
- Finn BE, Forsén S. . Structure. January 1995, 3 (1): 7–11. PMID 7743133. doi:10.1016/S0969-2126(01)00130-7.
- Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. . Cell. October 2008, 135 (1): 110–22. PMID 18854159. doi:10.1016/j.cell.2008.08.006.
- Nelson MR, Thulin E, Fagan PA, Forsén S, Chazin WJ. . Protein Sci. February 2002, 11 (2): 198–205. PMC 2373453. PMID 11790829. doi:10.1110/ps.33302.
外部链接
- Nelson M, Chazin W. . Vanderbilt University. [2009-08-29]. (原始内容存档于2009-08-25).
- Haiech J. . European Calcium Society and the Université Libre de Bruxelles. [2009-08-29]. (原始内容存档于2009-08-09).
upon request to haiech@pharma.u-strasbg.fr
- Yang J. . Georgia State University. [2009-08-29]. (原始内容存档于2009-10-12).
prediction server for EF-hand calcium binding proteins
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.
